About MICAS3.0 & MICdb3.0
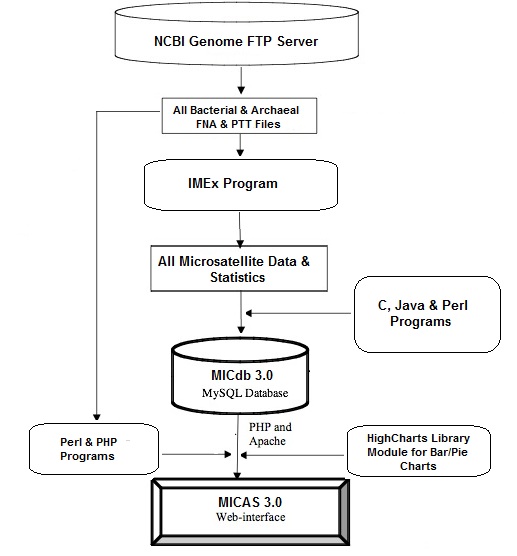
MICAS3.0 is an interactive user-friendly web-based analysis server to find non-redundant microsatellites of a selected bacterial or archeal genome sequence. MICAS has been connected to the database MICdb3.0 that hosts microsatellites extracted from more than 4000 published prokaryotic sequences. The extraction of microsatellites is done using the software tool Imperfect Microsatellite Extractor (IMEx) (Suresh Mudunuri and Nagarajaram HA, Bioinformatics, 2007) and have been compiled into a relational database.
Database Construction
All the sequenced Bacteria and Archaea genomes have been downloaded from the NCBI ftp server and have been submitted to IMEx. All perfect repeats with a minimum tract length of 6bp have been extracted. The parameters used for IMEx are as follows: Repeat Type: Perfect; Min. Repeat Number: mono:6, di: 3, tri-hexa:2). The output files of IMEx have been parsed using C & JAVA programs and the data has been inserted into the tables of MICdb database. The database has been constructed using MySQL.
Web-Interface Construction
MICdb3.0 has been connected to a user-friendly web-interface named MICAS3.0 (Microsatellite Analysis Server) to facilitate the users in accessing the data from MICdb and to analyse the microsatellites and their distribution at genome level. The web interface has been developed using HTML and CSS. The server side scripting has been done using PHP and AJAX. The high quality graphs and charts used by MICAS to visualize the query results have been generated using advanced JavaScript library named HighCharts. MICAS and MICdb have been hosted on a Linux Server containing Apache web-server and special care has been taken to ensure the interactivity and user-friendliness of the system. The earlier version of MICAS (Sreenu et. al., 2003) had 3 different retrieval methods - Classic, ORF based search and Advanced Search which have been modified and enhanced for user convenience and ease of access of data. The current version MICAS3.0 has 3 unique and different data access modules - Browse (search by alphabetical order of genomes), Advanced Search (search by user criteria) and Pair-wise Comparison of Genomes (compare genomes for microsatellite distribution).
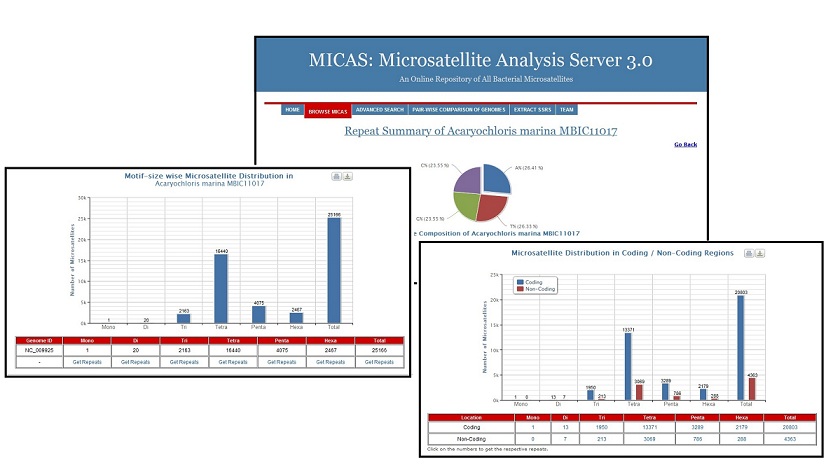
Genome Browse Module
Using the Browse module of MICAS, the user can easily browse through all the genomes of bacteria and archaea arranged in alphabetical order. On clicking a particular genome, the complete summary information of the genome related to nucleotide composition, microsatellite distribution in coding / non-coding regions,

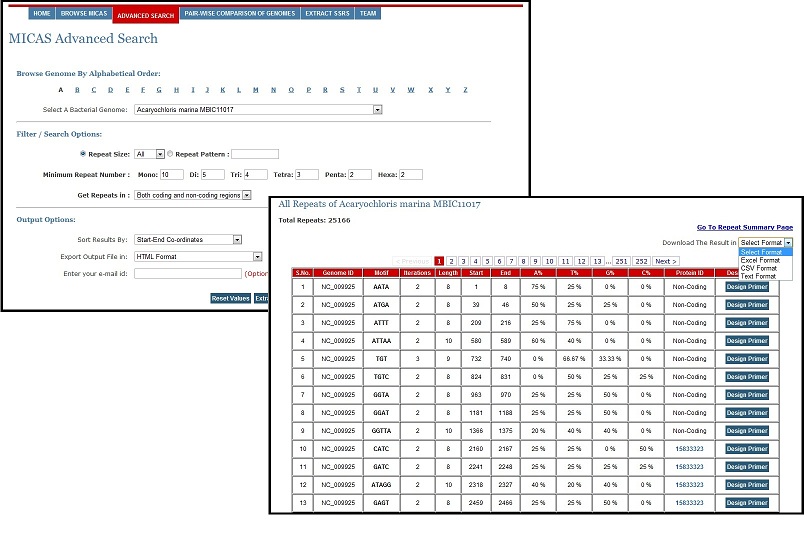
Advanced Search Module
To facilitate users for getting repeats based on specific search criteria, MICAS3.0 has been provided with an Advanced Search module. Using the Advanced Search module, users can select a particular genome of interest and also specify his/her search criteria and filter repeats results accordingly. Advanced Search module can filter repeats of a particular size (mono, tri, tetra etc.,); can get repeats of a particular pattern (CAG, Poly A etc.); can set the minimum repeat number of each motif size; and can filter repeats of only coding or non-coding regions. Moreover, the Advanced Search module allows you to define the output format (HTML, Excel, CSV or Text) and also sort results based on motif, motif-size, or tract length.
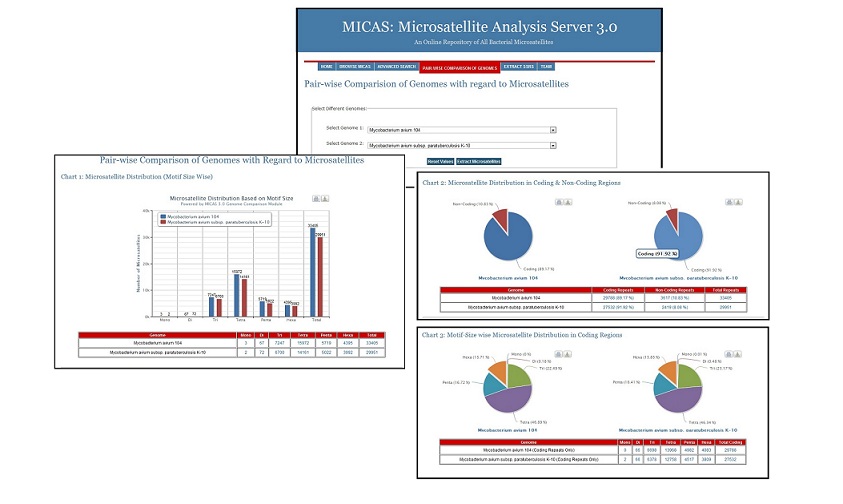
Genome Comparison Module
Using this module, the users can select 2 different genomes of interest and compare the microsatellite distribution in both the genomes. Microsatellite information pertaining to the distribution based on motif-size, distribution in coding/non-coding regions, motif-size wise distribution & the density of repeats in coding regions of both the genomes can be generated. The results are also displayed neatly in the form of bar/pie charts as well as in tabular formats.