About ChloroMitoSSRDB 2.0
Chloroplast represents an important autotrophic organelle, which is involved in several important functions, of which the most important is photosynthesis. All plastids studied to date contain their own DNA, which is actually a reduced "genome" derived from a cyanobacterial ancestor that was captured early in the evolution of the eukaryotic cell. Land plant chloroplast genomes typically contain around 110-120 unique genes. Some algae have retained a large chloroplast genome with more than 200 genes, while the plastid genomes from non-photosynthetic organisms may retain only a few dozen genes (Cui et al. 2006). Several chloroplast genes (rbcL, matK, ycf, psbA) have been successfully used as barcodes to delineate the evolutionary biology among the plant clades. Mitochondrial genomes, present a rapidly a wide array of rapidly evolving genomes in sequence context and genome content (Gray et al., 1999). Relatively high incidence of RNA editing and trans-splicing of coding sequences (Hiesel et al. 1989; Knoop 2004) has been observed in mitochondrial genomes.
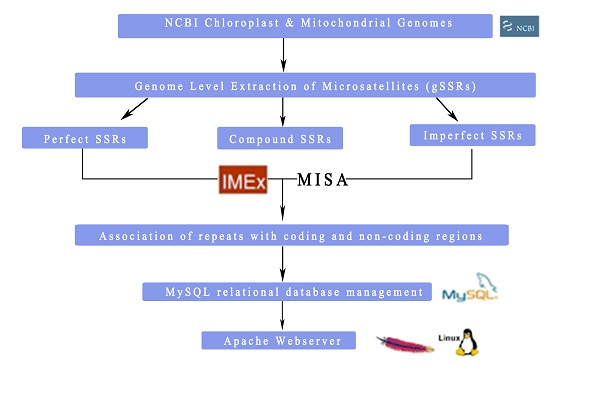
ChloroMitoSSRDB 2.00 database cum webserver gives systematic and curated information on the presence of microsatellites in the so far available chloroplast and mitochondrial genomes along with the primer pair information. Additionally, it allows the users to interact and visualize the microsatellite association with the coding and non-coding regions. The present version is a comprehensive and on-line portal, which unifies the mining of the SSRs and the primer pairs from the first and next generation sequencing (NGS) datasets from the organelle genomes. For the construction of the ChloroMitoSSRBD 2.00, all the chloroplast and the mitochondrial genomes have been scanned for the presence of the perfect, imperfect and compound repeats. The unified web-interface allows the user to select the chloroplast and mitochondrial genomes and also the SSRs searches from the IMEx and MISA respectively. The repeat statistics interface gives the user, the information on several repeat statistics including the repeat unit and length, number of repetitions of the repeat unit and position of the repeat in the genome and also its presence in the genic or the intergenic regions. Additionally, in MISA it also gives the information on the primer pairs along with the repeat annotation. Advanced search option allows the users to select the repeat type and can view its repeat statistics in the chloroplast and mitochondrial genomes.
The webserver section allows the users to select the SSR/repetitive pattern search using three embedded algorithm viz. IMEx, MISA and Reputer for whole genome or gene based searches.
Additionally, it also allows users to upload the NGS reads of the chloroplast and mitochondrial genomes to analyze and identify the SSRs from the NGS reads along with the primer pair information.
Citation:
Gaurav Sablok, Suresh B. Mudunuri, Sujan Patnana, Martina Popova, Mario A. Fares, and Nicola La Porta (2013) ChloroMitoSSRDB 2.0: Open source repository of perfect and imperfect repeats in organelle genomes for evolutionary genomics. DNA Research, 10.1093/dnares/dss038. (Impact Factor: 5.1)
We would also appreciate suggestions and useful constructive point for the future upgrades and release of the database.
References:
- Gray MW, Burger G, Lang BF (1999) Mitochondrial Evolution. Science 283: 1476-1481.
- Cui L, Veeraraghavan N, Richter A, Wall K, Jansen RK, Leebens-Mack J, Makalowska I, dePamphilis CW (2006) ChloroplastDB: the chloroplast genome database. Nucleic Acids Research 34: D692-696.
- Hiesel R, Wissinger B, Schuster W, Brennicke A. 1989. RNA editing in plant mitochondria. Science 246:1632-1634.
- Knoop V. 2004. The mitochondrial DNA of land plants: peculiarities in phylogenetic perspective. Curr Genet. 46:123-139.
ChloroMitoSSRDB 2.0 Architecture